Bernie Devlin, PhD, on the Autism Sequencing Consortium’s Large-Scale Genetic Study of the Neurobiology of ASD Published in Cell
Bernie Devlin, PhD, Professor of Psychiatry and Clinical and Translational Science, earned his PhD in botany, conducting his early research in plant biology. His postdoctoral research in genetics in plant populations ignited a career-long love for statistical modeling, and today he is an internationally recognized expert in building statistical models to understand the genetic epidemiology of simple and complex diseases.
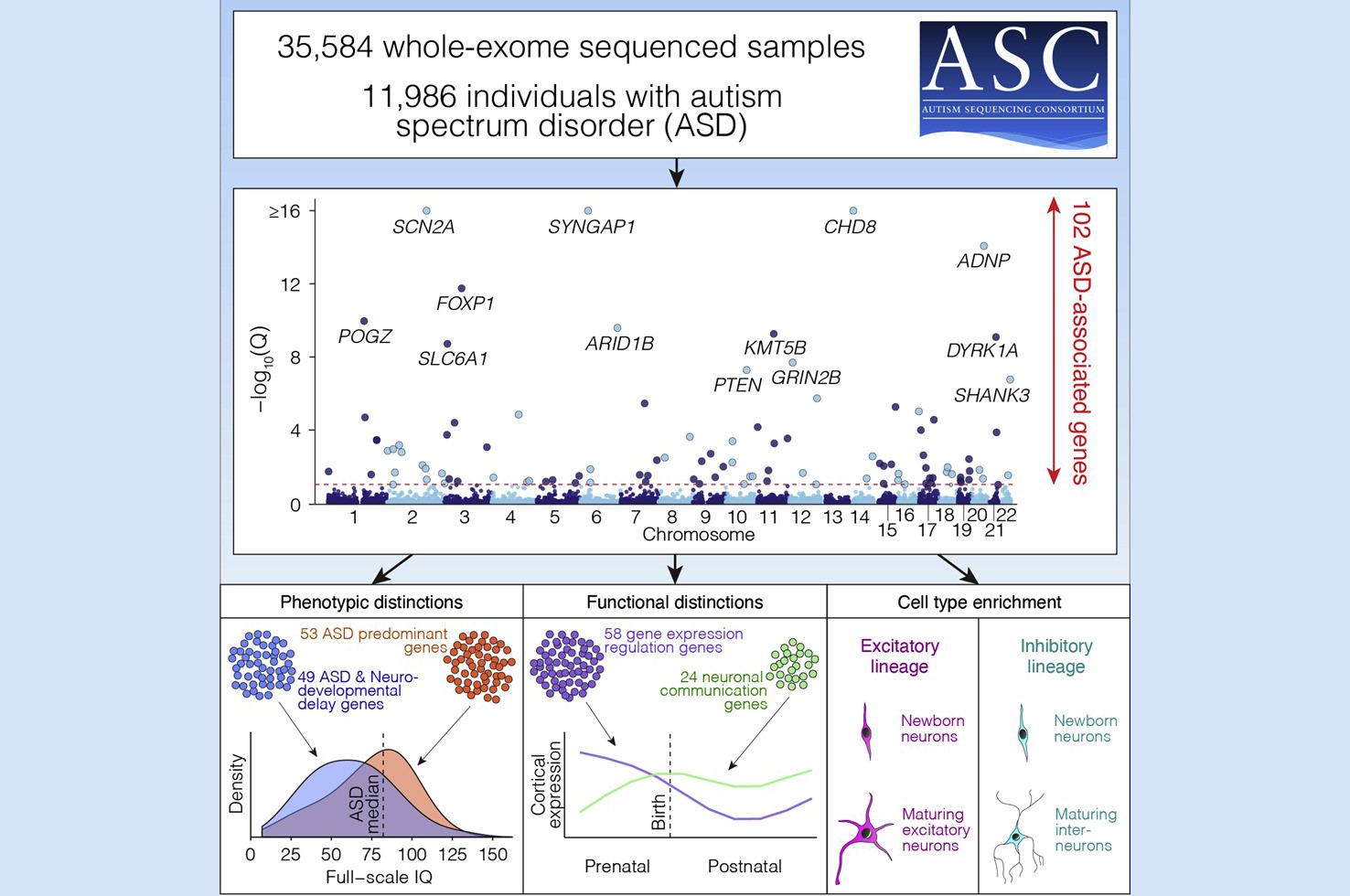
Dr. Devlin is one of six principal investigators for the National Institutes of Health-funded Autism Sequencing Consortium (ASC), a large-scale international genomic consortium that shares autism spectrum disorder (ASD) samples and genetic data. In a recent study published in the journal Cell, the ASC analyzed 35,584 unique human samples with the goal of identifying genes that, when disrupted by mutation, have a substantial impact on the risk for autism. Dr. Devlin, in collaboration with several colleagues, ensured sound and effective statistical analysis of the genetic data provided to or produced by the study.
The ASC began with the key observation that young people with autism carry mutations that are more frequently occurring—and more damaging—than could be attributed to chance. The research was guided by two important questions, explained Dr. Devlin: “What do we expect by chance? Furthermore, when the mutational pattern in genes deviates from what we expect by chance, how much of a deviation is enough to implicate the gene in risk?”
The ASC developed a list of 102 genes that are implicated in the risk for ASD. This genetics-first approach to investigating autism stems from the understanding that while all psychiatric disorders result from altered neurobiology, the altered neurobiology of autism is subtle. Because most if not all of the identified genes impact neurodevelopment beyond ASD, one of the scientists’ initial challenges was determining how to tease apart the list to learn which genes, when mutated, affect development broadly and which affect autism more prominently. Moreover, “autism is a complex genetic trait, and implicating single genes won’t be immensely helpful for understanding its neurobiology,” explained Dr. Devlin.
After developing the list of ASD-implicated genes, the investigators compared the mutational patterns in the autism cohort with mutational patterns observed in individuals who participated in a study pertaining to developmental delay and intellectual disability (with or without autism). Recognizing that substantial overlap would occur between the two studies, the ASC separated the genes into those genes that carried relatively more mutations in autism cohort and those genes that carried more mutations in the developmental disability cohort. The hope was that this simple classifier, which separated genes into two bins, would tend to separate genes into those affecting development broadly and those affecting autism more prominently. In analyzing ASD probands, they demonstrated that these two classes of genes did, on average, predict features of development consistent with the classification. When genes categorized as having “profound impact on development” were mutated, the ASD carriers of these mutations had lower IQ and walked later than did ASD carriers of mutations in genes falling in the ASD-primary category. “We didn’t make any claims that we could separate these genes like Moses parted the Red Sea, but we were getting somewhere. I think we are going to be able to start teasing out what genes, when mutated, cause more profound impact on certain phenotypes than others.”
To address the second challenge, related to the genetic complexity of ASD, the team investigated when and in what cell types ASD genes are first expressed. They analyzed gene expression patterns in fetal cells of a variety of types from brains at different developmental stages, data that were gathered from a previous study. They found that many ASD risk genes were first expressed in neuroprogenitor cells, which develop into either excitatory or inhibitory neurons, among other types of cells, and the vast majority were expressed in both excitatory and inhibitory neurons by 24 weeks post conception.
“These results do not lend themselves to a simple story for ASD,” said Dr. Devlin. “What we hoped would demystify the neurobiology of ASD has suggested instead that there is a reason why neurobiologists have not seen any obvious and consistent features in ASD brains. There appear to be multiple ways to alter the neurodevelopment of the brain to produce the social communication and behavioral deficits recognized in ASD.”
Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism
Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An JY, Peng M, Collins R, Grove J, Klei L, Stevens C, Reichert J, Mulhern MS, Artomov M, Gerges S, Sheppard B, Xu X, Bhaduri A, Norman U, Brand H, Schwartz G, Nguyen R, Guerrero EE, Dias C; Autism Sequencing Consortium; iPSYCH-Broad Consortium, Betancur C, Cook EH, Gallagher L, Gill M, Sutcliffe JS, Thurm A, Zwick ME, Børglum AD, State MW, Cicek AE, Talkowski ME, Cutler DJ, Devlin B, Sanders SJ, Roeder K, Daly MJ, Buxbaum JD.
Cell. 2020 Feb 6;180(3):568-584.e23. doi: 10.1016/j.cell.2019.12.036. Epub 2020 Jan 23.
